qPCR plays a vital role in addressing today’s healthcare questions, with applications ranging from gene expression analysis to mutagenesis studies. The revolutionary technique amplifies DNA in samples, thereby enabling scientists to accurately measure minute concentrations of analytes. However, qPCR’s biggest strength also leads to its biggest challenge: any contamination in the sample, no matter how small, is also amplified.
Contamination can significantly compromise sample integrity in qPCR experiments. Crucially, it can lead to either false negatives or false positives in a sample, severely impacting critical research and analysis and rendering your results unreliable. What’s more, finding the source of contamination takes time, and the necessary experimental repeats are expensive, making projects less viable.
So, how can you ensure the integrity of your qPCR samples? This article explores where contamination arises from, and how to identify it. To improve sample integrity, we highlight approaches you can take to minimize contamination
Contamination in qPCR Experiments
Where Does Contamination Come From?
The causes of contamination in qPCR experiments fall broadly into two main categories: environmental contamination, and chemical contamination.
Environmental contamination refers to any contaminant introduced by the handling of the experiment. As with most experiments, instruments and devices can expose samples to contamination, including lab equipment and liquid handling devices such as pipettes and pipette tips. But for qPCR experiments, amplification carryover contamination poses an additional and significant threat to sample integrity. This is caused by the formation of aerosols that occurs when opening a tube or a plate, meaning some sample can escape into the environment and rapidly amplify. Most notably, amplicons can be released, leading to false positives.
Chemical contamination can occur from any of the chemicals used within the laboratory. Components such as buffers, reagents, and nucleotides can all potentially bring contamination to your qPCR experiment: therefore, you should use only the highest quality and purity chemicals.
How to Identify if Your qPCR Experiment is Contaminated
Ensuring you have proper controls in your qPCR assay is the key to identifying contamination early, thereby minimizing disruption to your experiment. Appropriate controls also enable troubleshooting, optimization of conditions, and validation of the experiment and subsequent results. But what types of control should you use?
No Template Controls (NTC)
NTCs are the negative, or the blank control, for the qPCR experiment. The reaction set-up should include NTC wells containing all the reaction components except the DNA template. In the absence of contamination, no amplification is expected during the thermocycling. If any amplification is observed, there is likely contamination from either a reaction component or the environment.
Negative Control of Extraction (NCE)
Similar toNTCs, NCEs are the negative control for the extraction process. These can enable the identification of contamination that occurs in the upstream process to provide the DNA required in the qPCR experiment.
No Amplification Controls (NAC)
In NACs, DNA polymerase is omitted from the PCR reaction. NACs act as a control for background fluorescence that is not a function of this reaction.
Positive Controls (PC)
Two types of PCs are common: exogenous and endogenous. The first, exogenous PCs, check if the reaction is working, and confirm that any negative results are true negatives. Here, an unrelated sequence —often a genome from another species —is spiked into the samples. If there is no amplification, it indicates that your PCR is not properly set, or your sample contains an inhibitor. The latter, endogenous PCs, are used to normalize the samples, and alsoensure the reaction is working. For these PCs, reference genes are commonly used.
Resources & Solutions for your PCR / qPCR Applications
PCR has enabled many applications in fields such as diagnostics, forensics, and research. A single copy of DNA can be exponentially amplified by a series of cycles of temperature changes. The process can be contaminated by a single copy of an undesired DNA sequence, and special tools are often needed. Gilson has been involved in the area of PCR for many years and has a range of products to assist in this process.
Explore Now
Avoid Contamination in Your qPCR Experiments
Once contamination has been introduced into a qPCR experiment, you cannot remove it. It is, therefore, crucial to prevent contamination of the sample in the first place. To avoid contamination and ensure confidence in your sample integrity, there are multiple approaches you can consider, such as separating your spaces, undertaking clean sample processing, and introducing automation. We explore each of these in more detail below.
Separate Your Spaces
One of the most effective approaches to prevent contamination is to separate your spaces, ensuring you have dedicated areas for different parts of the workflow. The benefits of this solution can be limited if there is insufficient lab space, but at a minimum, you should create separate areas for pre-and post-amplification areas. If space allows, you can further secure sample integrity by installing a third area for combining sample dilutions and reaction mix. And ideally, the room’s ventilation systems should not be linked, reducing the chances of amplification carryover contamination.
Once you have separated the spaces, fine-tune the environments and workflows to protect your samples further. The specific areas should have their own separate equipment: pipettes, pipette tips, and PPE can all facilitate contamination. To avoid cross-contamination between reaction stages, personnel should work linearly where possible. Crucially, you should not enter the pre-amplification area after working in the post-amplification area.
Finally, remove any other sources of contamination. Make sure to frequently decontaminate any work surfaces and equipment using a 10% bleach solution. And remember, you can be a source of contamination, too. Before entering a pre-amplification area, ensure you decontaminate all your belongings: even items such as jewelry can pose a contamination risk.
Use the Right Liquid Handling Tools
Once you’ve set the foundations with separate spaces, it’s time to pick the right equipment. Manual liquid handling runs a higher risk for contamination, particularly due to aerosols, and you should reduce risks where possible. Make sure to carefully select both your pipettes and pipette tips, for example, as they can act as contamination sources.
When selecting a pipette, it is essential that you can autoclave it to remove any contamination. Note that not all parts of your pipette may be autoclavable, so refer to the manufacturer’s instructions to determine the most suitable protocol.
If you are using air displacement pipettes, filter tips offer effective aerosol protection to keep your pipette and sample free of contamination. Ensure the filters and the pipette tips are made using high quality, durable materials such as polypropylene. In addition, look for those that were manufactured in a sterile, clean-room environment to ensure they are contaminant-free.
Depending on the experiment need, you can also use positive displacement pipettes if appropriate. These pipettes are compatible with a capillary piston (CP) pipette tip, and as there is no air between the piston and the reagents, it reduces contamination risk.
Consider Adopting Automation
If you want to reduce contamination risk further, consider implementing an automated solution. While you can effectively minimize contamination during the liquid handling steps, manual processes in qPCR experiments inherently bring additional risks of contamination and error being introduced, such as adding the wrong sample to the wrong well. You can overcome this by automating processes such as liquid handling to minimize human intervention, and therefore eliminate the risk of accidental contamination.
You can also experience additional sample integrity beyond contamination avoidance when automating your processes. It can, for example, enable the normalization of both RNA and DNA concentrations, as recommended by Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines (Figure 1). Variable RNA and DNA amounts in the experiment lead to differences in the efficiency of reverse transcription reactions and in quantification cycle values. Automate this procedure to remove the risk of error and user-to-user variability and thereby improve your sample reliability.
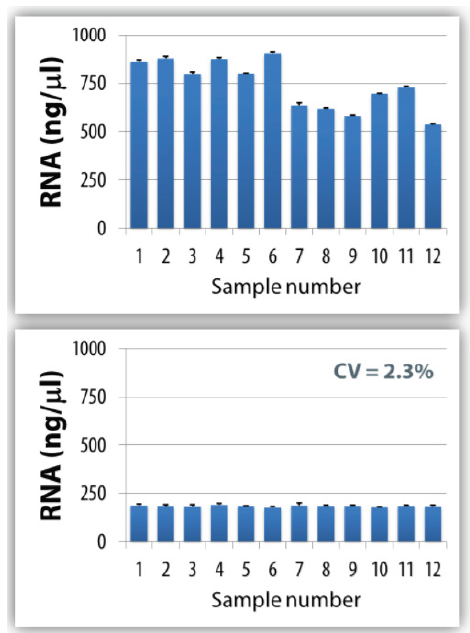
Figure 1. Human RNA sample normalization using PIPETMAX® Normalization Assistant, a software wizard that performs the required calculations from imported DNA concentrations. The graphs show the concentration of each sample before (top) and after (bottom) normalization. Error bars show the range for Nanodrop concentration measurements.
Contamination in your qPCR workflow can compromise confidence in your experimental results and should be avoided at all costs. Including the right controls from the start of your experiment can help you identify contamination before it’s too late —saving time and money. Additionally, separating your spaces, using the right liquid handling tools, and adopting automation can give you the greatest chance of avoiding contamination in your qPCR experiments and upholding sample integrity.
With over 50 years of experience, Gilson has the knowledge, expertise, and equipment to support the scientific community in maintaining complete confidence in sample integrity. Visit our learning hub to discover more about improving sample integrity in your qPCR experiments.
PIPETMAX® Automated Pipetting System
PIPETMAX® is an automated pipetting solution for the efficient processing of high-throughput biological assays. It will help you to improve on the accuracy, reproducibility, and consistency among all samples processed. Unlike traditional automation platforms, PIPETMAX comes in a benchtop size that easily fits into any lab.
Request a Quote